Phylogenetics is the study of evolutionary changes and the search for answers to different evolutionary questions such as how a speciation event occurred, and why. I am super excited about this topic and the things that I have been doing in the lab and that I have been reading, and hope this blog will be interesting for everyone else to read.
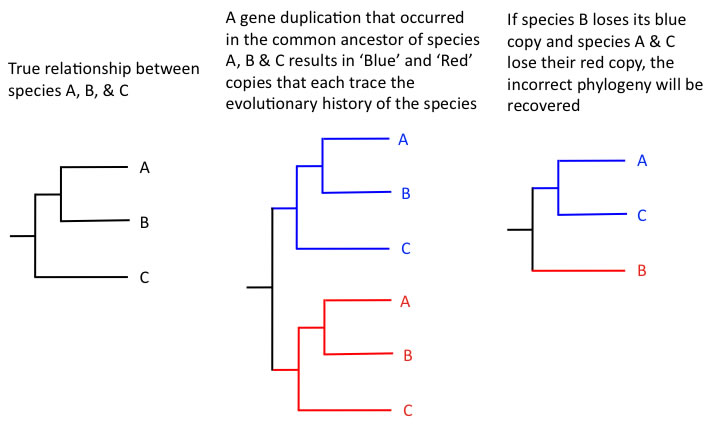 |
| Fig. 1: Paralogy vs. Orthology |
So, basically, over the year, I have been given several articles to read and two books that explain phylogenetic methods and important concepts. One of the main things that I kept bumping into when I was reading the articles about the lupine research (which are the hardest things to understand; you literally have to break the whole article into thousands of pieces and define every third word) are the paralogy, orthology, and coalescence theories. These three describe different approaches to analyzing a phylogenetic tree. Paralogy is not favored when creating a phylogenetic tree, since it is the creation of a phylogenetic tree solely by analyzing gene sequences. Though under first thought, this may seem more accurate, it actually isn’t: analyzing only gene sequences may lead the observer to indiscernible gene duplication events because we cannot find out if the gene duplication event occurred or not, or gene shutdown events, which may dramatically alter the observed phylogenetic tree for the specific species from the theoretical true phylogenetic tree (Fig. 1) However, this does not mean that we should completely throw away gene sequencing from phylogenetic analysis; this would be unproductive in terms of the advances in phylogenetics in the last century.
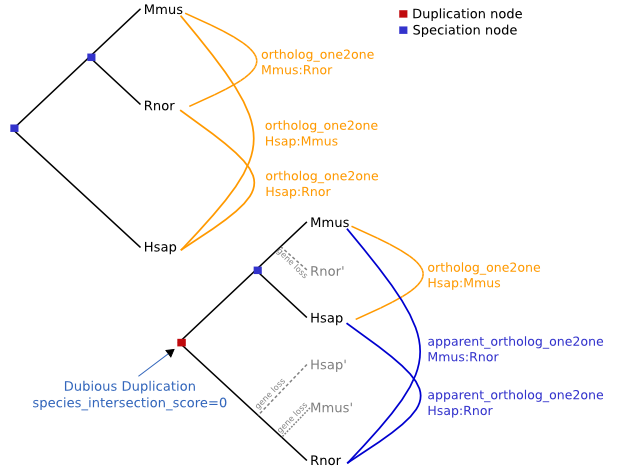 |
| Fig. 2: Speciation vs. Gene duplication event |
Orthology, on the other hand, observes the speciation events instead of solely the genetic events when creating a phylogenetic tree. This method is much more favored by systematists than paralogy, since it decreases the probability of errors that could be made compared to the theoretical phylogenetic tree of the species. A speciation event does not look at genetical changes in the sequences of different species in the same genus; rather it looks at different biogeographical events, fossil evidence, etc. in addition to gene sequences to make sure that the conclusions are more accurate (Fig. 2).
Coalescence theory, to be frank, is quite difficult to grasp; I still have difficulties understanding the concept when reading articles and books. In general, it is a way to trace back speciation lineages through time (by the “trunk” of the tree) to the present time to see how closely or just how different species are related. When coalescence theory is depicted in books and articles, it is usually shown as an inverted tree, which allows the reader to trace back in time instead from the past to the future, as a normal tree allows one to do (Fig. 3).
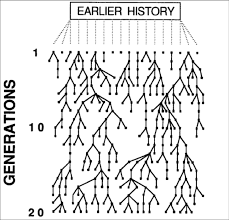 |
| Fig. 3: Upside down tree depicting coalescence theory |
Another topic that keeps popping up in my reading is ladder thinking versus tree thinking; Lamarck vs Darwin. Many people long ago believed that evolution occurs in a ladder model; this was also the preference of Lamarck, a biologist who believed that, for instance, if a giraffe wanted to reach some food on a tall branch, all it had to do was stretch really hard, and its neck would become longer, and that when it would reproduce, this trait would simply be passed on to its offspring. Darwin, on the other hand, after his trip to the Galapagos, differed. He believed that evolution occurred in a tree model.
The main difference between a ladder model and a tree model is the fact that a ladder model allows the observer to compare two living species as one being the ancestor of the other, when really it is not possible for us to develop from currently living species. We are not related to monkeys; we are related to now extinct apes that lived long ago, speciated at some time into neanderthals, which became extinct but speciated into homo-sapiens. That is the true model of evolution.
Currently, I’ve been spending nearly two hours per week in the lab working on my lupine samples from Peru. The original plan was that I would quickly sequence the plants, upload the information into genbank, and be done and ready to continue on to “real” research. Unfortunately, lab science is not that easy. We had to redo the PCR reaction (polymerase chain reaction) three times before it finally worked (by changing the buffer type and allowing closer primers to be together). Only this week were we finally able to purify the DNA, and next week we will sequence them and be able to see if they really are related (really, all of this is just an exercise; nothing extraordinary or new). Afterwards, Dr. Tank has found a project that he would like me to work on; plants from the Galapagos that may have a similar evolutionary pattern to that of the Galapagos finches that Darwin found. (Though nobody believed that plants are interesting, no one has really looked into that hypothesis yet). I am super excited to do this project, and can’t wait to find out about the results. Lab work is a lot more fun for me than most people may think; yes, it can be a bit monotonous when I am doing the same protocol over and over, but I think that it is a great opportunity to be able to explore real science in the lab.
So… this is what I have been doing lately. Hope this was interesting!
~Valeria